Tiling stacks in ImageJ
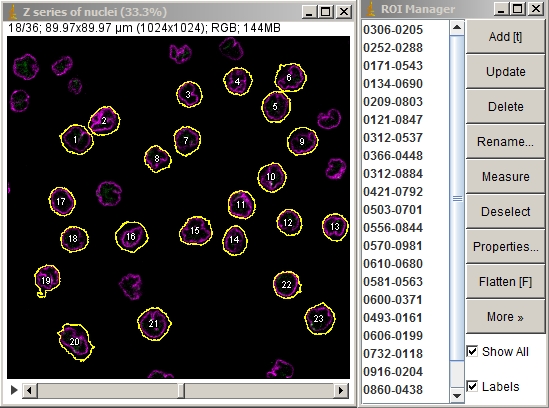
This page describes marking a field of isolated cells in a Z series such that each cell is individually made into a 3D movie and the movies are montaged into a single stack. Alternatively, the marked cells themselves may be extracted and montaged as a stack without the 3D processing.
The goal was to visualize and measue the distance of the FISH probes relative to the nuclear membrane.
The nucleus was labeled by immunofluorescence for Lamin A shown here in magenta. The FISH probes are shown here in green. Imaging with a 63X NA 1.4 planapochromat lens with a Zeiss LSM 710 laser scanning confocal.

To export each cell as its own individual Z series, run "clip all ROIs in Roi Manager each as a stack [F4]" in crop_and_montage_stacks_v100.txt (written circa 2011, perhaps needs rewriting) or to make a 3D rotation of each cell as pictured below, run "pull all Z series from two windows, merge, and 3D" in quick_3D_macro_v100.txt with the appropriate window names inserted into the macro.
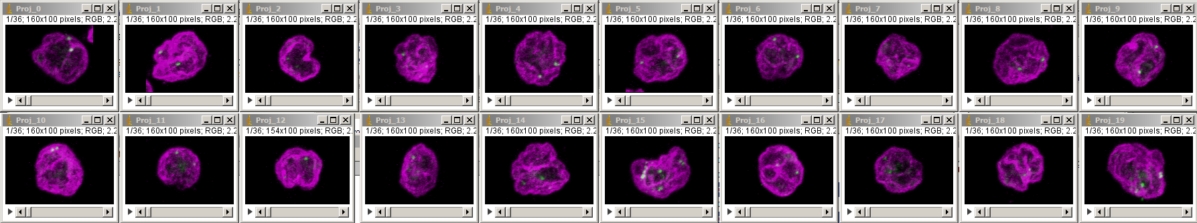
Run the macro "combineStacks" in crop_and_montage_stacks_v100.txt
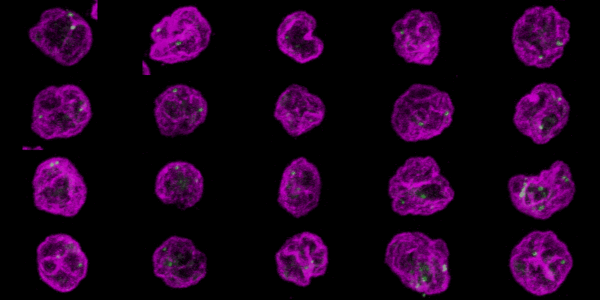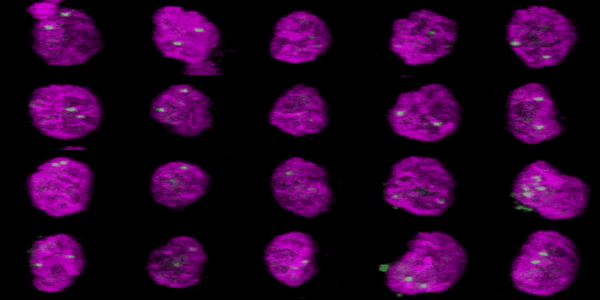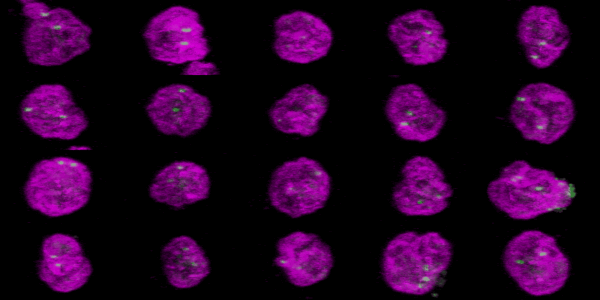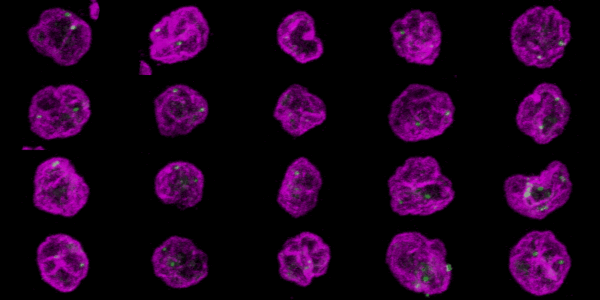
Click here for the resulting stack.
comments, questions, suggestions: Michael.Cammer@med.nyu.edu or mcammer@gmail.com